Joint Research Group Macromolecular Crystallography
HZB Fragment Screening Facility

Fragment screening
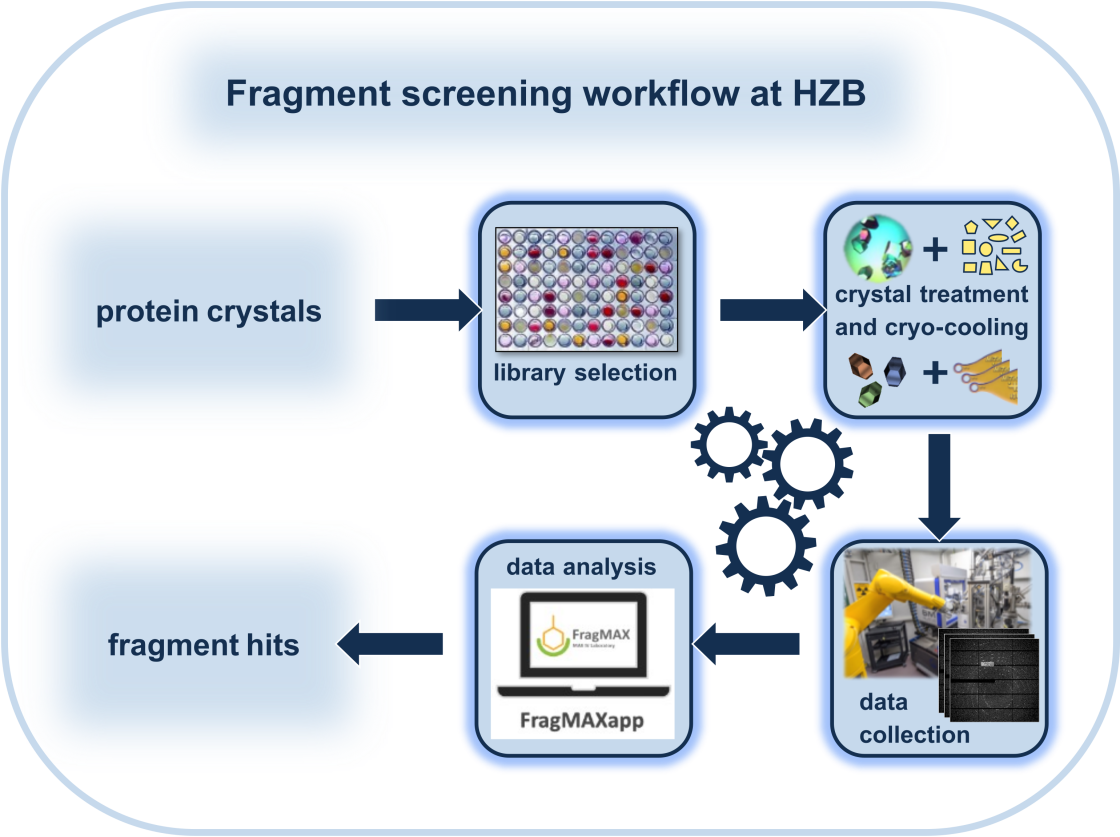
Fragment screening (FS) is a popular approach to identify the binding of small organic molecules (fragments) to protein targets. This may be done to elucidate the function of a protein or to identify chemical structures, which may be developed into tool compounds or leads. Probably the most suitable method for FS is X-ray crystallography. Crystallography is not only able to identify very weakly binding fragments (with binding constants in the mM range), it also provides information on where and how the fragments bind to the protein. Based on the obtained fragment hits, commercially available compounds can be used to find follow-up molecules that bind with higher potency, thus initiating a tool compound or lead development endavor.
At the HZB, we have developed a fragment-screening workflow (see below) and assembled several libraries, which are available to our users. In addition, expert assistance to plan and conduct preliminary test to optimize the soaking condition as well as the final experiment will be provided. As part of our workflow, we support the largely automated analysis of the data sets and first steps in hit advancement. In order to conduct a FS campaign, it is important to familiarize yourself with the workflow and the requirements to your crystal system. Get in touch with us if you would like to know more about the workflow or the feasibility of it for your target. (contacts: Dr. Melanie Oelker and Dr. Manfred Weiss.)
At present, a fragment-screening campaign using the HZB fragment libraries requires the signing of a collaboration contract. Beamtime proposals can be submitted through HZB's electronic user office GATE. Addditionally, international user experiments have the possibility to apply for funding through the EU Horizon 2020 Project iNEXT-Discovery. For further information about that please contact Dr. Manfred Weiss .
Detailed Informations

References
- Metz A, Wollenhaupt J, Glöckner S, Messini N, Huber S, Barthel T, Merabet A, Gerber HD, Heine A, Klebe G, Weiss MS (2021). Frag4Lead: growing crystallographic fragment hits by catalog using fragment-guided template docking. Acta Cryst. D, 77 (Pt 9):1168-1182
- Wollenhaupt J, Barthel T, Lima GMA, Metz A, Wallacher D, Jagudin E, Huschmann FU, Hauß T, Feiler CG, Gerlach M, Hellmig M, Förster R, Steffien M, Heine A, Klebe G, Mueller U & Weiss MS (2021). Workflow and Tools for Crystallographic Fragment Screening at the Helmholtz-Zentrum Berlin. J. Vis. Exp. 62208
- Barthel, T., Huschmann, F.U., Wallacher, D., Feiler, C.G., Klebe, G., Weiss, M.S. & Wollenhaupt, J. (2021), Facilitated crystal handling using a simple device for evaporation reduction in microtiter plates, J. Appl. Cryst.
- Wollenhaupt, J., Metz, A., Barthel, T., Lima, G. M. A, Heine, A., Mueller, U., Klebe, G. & Manfred S. Weiss, M.S. (2020), F2X Universal and F2X Entry: structurally diverse compound libraries for crystallographic fragment screening, Structure
- Schiebel, J., Krimmer, S. G., Röwer, K., Knörlein, A., Wang, X., Park, A. Y., Stieler, M., Ehrmann, F. R., Fu, K., Radeva, N., Krug, M., Huschmann, F. U., Glöckner, S., Weiss, M. S., Mueller, U., Klebe, G. & Heine, A. (2016). High-Throughput Crystallography: Reliable and Efficient Identification of Fragment Hits. Structure 24, 1398–1409.
- Sparta, K., Krug, M., Heinemann, U., Mueller, U., Weiss, M. S. (2016). XDSAPP2.0 J. Appl. Cryst. 49, 1085-1092.
- Schiebel, J., Radeva, N., Krimmer, S. G., Wang, X., Stieler, M., Ehrmann, F. R., Fu, K., Metz, A., Huschmann, F. U., Weiss, M. S., Mueller, U., Heine, A. & Klebe, G. (2016). Six Biophysical Screening Methods Miss a Large Proportion of Crystallographically Discovered Fragment Hits: A Case Study. ACS Chem Biol. 11, 1693-1701.
- Huschmann, F. U., Linnik, J., Sparta, K., Ühlein, M., Wang, X., Metz, A., Schiebel, J., Heine, A., Klebe, G., Weiss, M. S. & Mueller, U. (2016). Structures of endothiapepsin-fragment complexes from crystallographic fragment screening using a novel, diverse and affordable 96-compound fragment library. Acta Cryst. F72, 346-355.
